The latter of those two packages, venneuler, makes the whole process so criminally easy that you could safely ignore gplots altogether. That is, unless you want the actual counts of each group plotted as above. That may be a job for post-R vector image editing but why do it by hand when you can automate it?
November 2015 update:
There's also the Vennerable package. It's described here and doesn't appear to be in CRAN but can be downloaded from R-Forge. Vennerable can handle all kinds of exotic n-group Venns, with a maximum n of nine.
Vennerable depends on graph, a package available through Bioconductor.
I haven't used it yet but will likely do so soon. Example output will show up here (further update: Vennerable also needs RBGL from Bioconductor. I ran into some unresolvable dependency issues while testing Vennerable so it will have to wait until I really need a nine group diagram, but that may indicate larger problems.)
February 2016 update:
Vennerable is now on Github. You'll need the devtools package to install it that way.
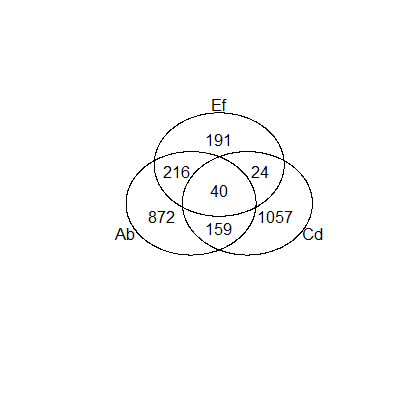
Vennerable has some neat features but lacks useful documentation. Here's a quick example.
Using the example data provided with the package, and cutting it down to just three groups:
data(StemCell)
VennRaw <- Venn(StemCell)
Venn3 <- VennRaw[, c("OCT4", "SOX2", "NANOG")]
plot(Venn3, doWeights = FALSE)and that gives you the basic diagram.